Breast Cancer Immunohistochemical Image Generation Challenge¶
Motivation¶
Breast cancer is a leading cause of death for women. Histopathological checking is a gold standard to identify breast cancer. To achieve this, the tumor materials are first made into hematoxylin and eosin (HE) stained slices (Figure 1). Then, the diagnosis is performed by pathologists by observing the HE slices under the microscope or analyzing the digitized whole slide images (WSI).
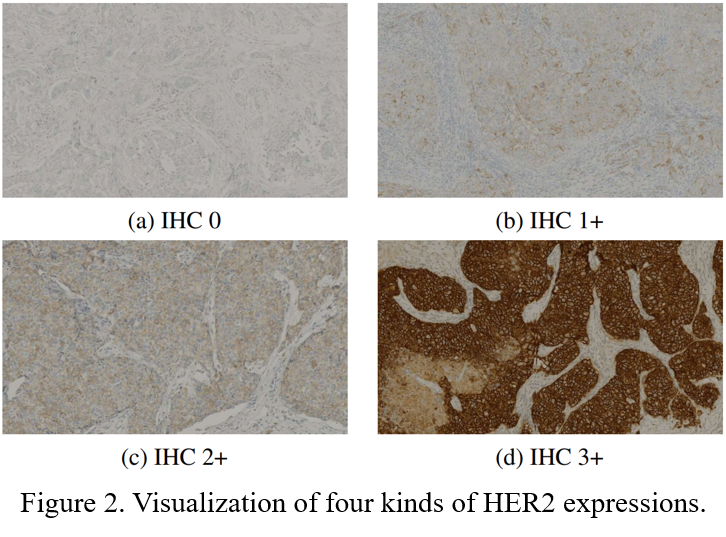
For diagnosed breast cancer, it is essential to formulate a precise treatment plan by checking the expression of specific proteins, such as human epidermal growth factor receptor 2 (HER2). The routine evaluation of HER2 expression is conducted with immunohistochemical techniques (IHC). An IHC-stained slice is shown in Figure 1. Intuitively, the higher the level of HER2 expression, the darker the color of the IHC image (Figure 2).

However, there are certain limitations in assessing the level of HER2 expression by IHC technology: 1)The preparation of IHC-stained slices is expensive. 2)Tumors are heterogeneous, however, IHC staining is usually performed on only one pathological slice in clinical applications, which may not entirely reflect the status of the tumor.
**Therefore, we hope to be able to directly generate IHC images based on HE images. ** In this way, we can save the cost of IHC staining, and can generate IHC images of multiple pathological tissues of the same patient to comprehensively assess HER2 expression levels.
Task¶
This is an image-to-image translation task that builds a mapping between two domains (HE and IHC). Given an HE image, the algorithm should predict the corresponding IHC image.
Baseline¶
To allow for a quick start with the challenge data and tasks, our team has released the code of Pyramidpix2pix, which can be seen as a baseline.
Dataset¶
We propose a breast cancer immunohistochemical (BCI) benchmark attempting to synthesize IHC data directly with the paired hematoxylin and eosin (HE) stained images. BCI dataset contains 9746 images (4873 pairs), 3896 pairs for train and 977 for test, covering a variety of HER2 expression levels. Some sample HE-IHC image pairs are shown in Figure 3.

Evaluation¶
We use Peak Signal to Noise Ratio (PSNR) and Structural Similarity (SSIM) as the evaluation indicators for the quality of the generated image. PSNR is based on the error between the corresponding pixels of two images and is the most widely used objective evaluation index. However, the evaluation result of PSNR may be different from the evaluation result of the Human Visual System (HVS). Therefore, we also use SSIM, which comprehensively measures the differences in image brightness, contrast, and structure.
final ranking = 0.6*SSIM ranking + 0.4*PSNR ranking
Submission¶
Participants need to submit IHC images generated based on HE images in the testset, the naming of each IHC image needs to be consistent with the corresponding HE image.
After the final ranking is released, high-ranking teams will be invited to submit a description of their methods (no more than 2 pages), including the method introduction, model structure and experimental settings, etc. We will co-submit to a top-ranking journal with top-ranking teams.
Timeline¶
July 2nd, 2022 (11:59 PM UTC 8 Beijing Time): Launch of challenge and release of train and validation data.
August 10th, 2022 (11:59 PM UTC 8 Beijing Time): Release of test data.
August 24th, 2022 (11:59 PM UTC 8 Beijing Time): Submission of test results begins.
October 7th, 2022 (11:59 PM UTC 8 Beijing Time): Deadline for submission of test results.
October 14th, 2022 (11:59 PM UTC 8 Beijing Time): Final ranking will be announced; Top-ranking teams submit their method descriptions. The final rankings are available here!
October 31st, 2022 (11:59 PM UTC 8 Beijing Time): Deadline for submission of method descriptions.
Notes¶
In order not to limit the performance of the participants, the use of publicly available external data (such as models pretrained on other publicly available datasets) is allowed. If you do this, you will need to declare what external data is used in the submission comments.
Citation¶
If you have used the BCI Dataset in your research, please cite the paper that introduced the BCI Dataset:
@InProceedings{Liu_2022_CVPR, author = {Liu, Shengjie and Zhu, Chuang and Xu, Feng and Jia, Xinyu and Shi, Zhongyue and Jin, Mulan}, title = {BCI: Breast Cancer Immunohistochemical Image Generation Through Pyramid Pix2pix}, booktitle = {Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) Workshops}, month = {June}, year = {2022}, pages = {1815-1824} }
Contact¶
If you encounter any problems please contact us directly by the following email:
